

# Install in R
install.packages("quickcode")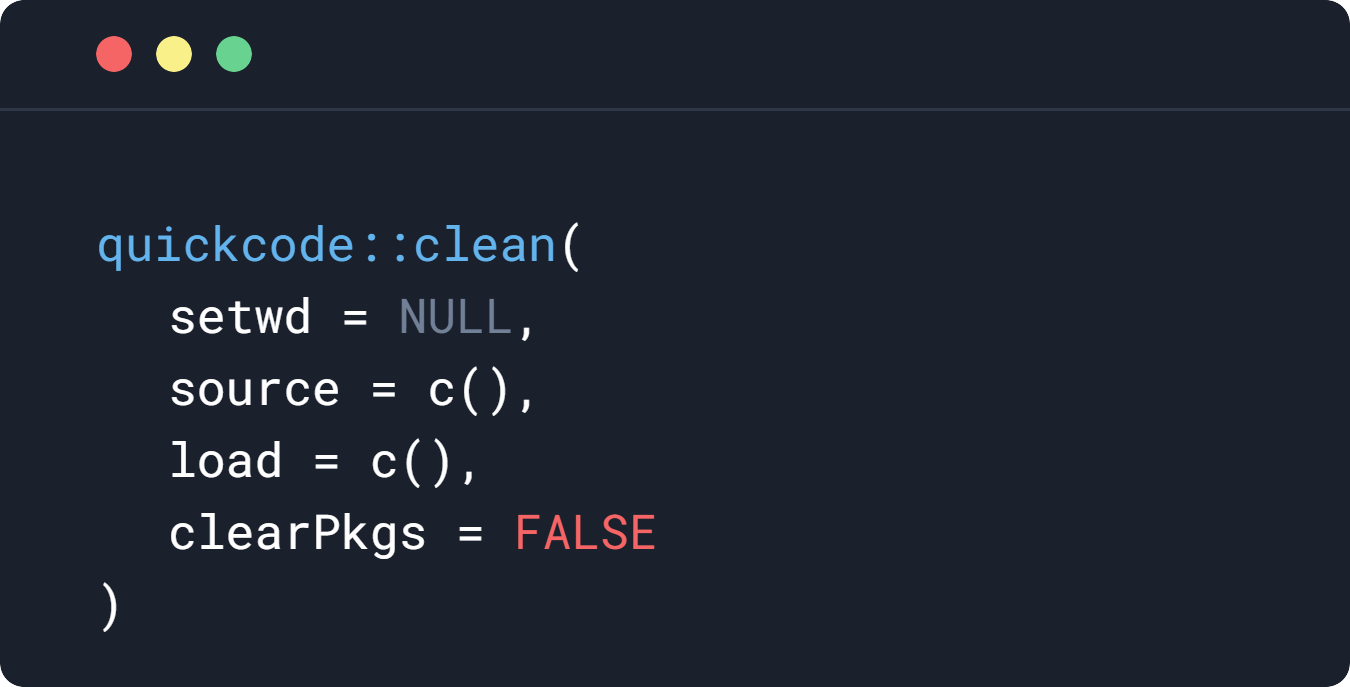


# Use the nullish coalescing operator using "or()" or "%or%"
ex.V1 <- 5
ex.V2 <- NA
ex.V3 <- NULL
ex.V4 <- ""
alternative <- 500
# Give an alternative result if the test is NULL NA or empty
or(ex.V1,alternative) # result will give 5 because ex.V1 is not NULL NA or empty
ex.V1 %or% alternative # result will give 5 because ex.V1 is not NULL NA or empty
ex.V2 %or% alternative # result will give 500 because ex.V2 is NA
ex.V3 %or% alternative # result will give 500 because ex.V3 is NULL
ex.V4 %or% alternative # result will give 500 because ex.V4 is empty
# Further chaining
ex.V2 %or% ex.V1 %or% alternative # result will be 5 because ex.V2 is NA but ex.V1 is 5
ex.V2 %or% ex.V3 %or% alternative # result will be 500 because ex.V2 is NA and ex.V3 is NULL
#load libraries and print names along with versions
quickcode::libraryAll(
dplyr,
r2resize,
ggplot2
)
#simple conversion between boolean types
#input type is "vector"
baba <- c(TRUE,"y","n","YES","yes",FALSE,"f","F","T","t")
as.boolean(baba,1) # return vector as Yes/No
as.boolean(baba,2) # return vector as TRUE/FALSE
as.boolean(baba,3) # return vector as 1/0
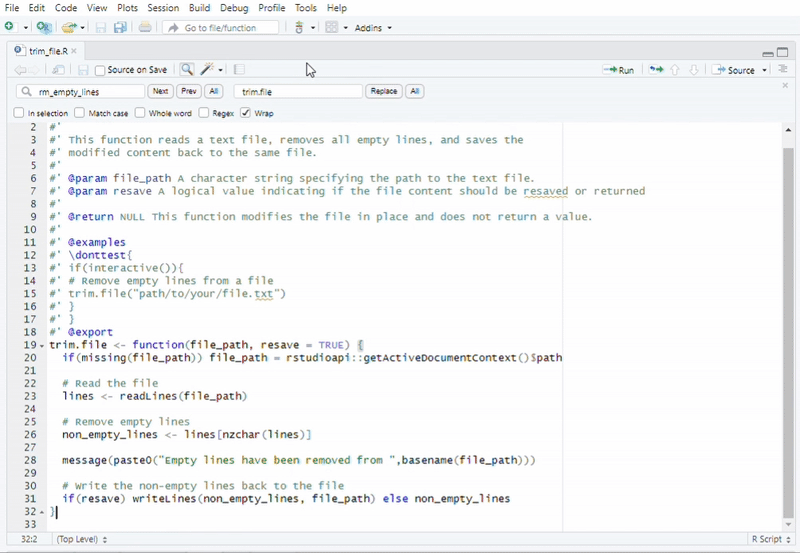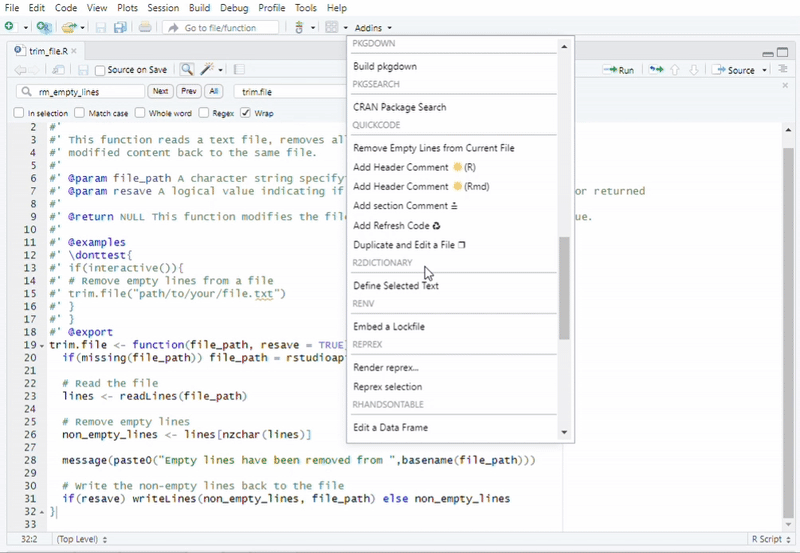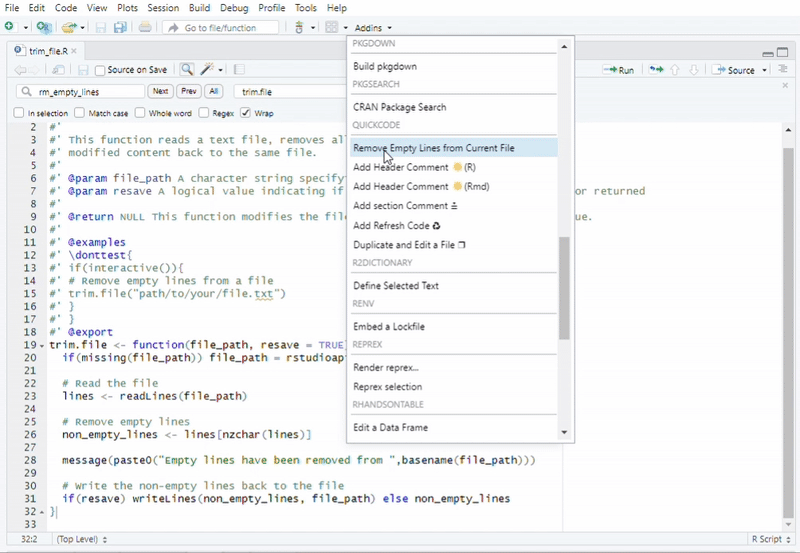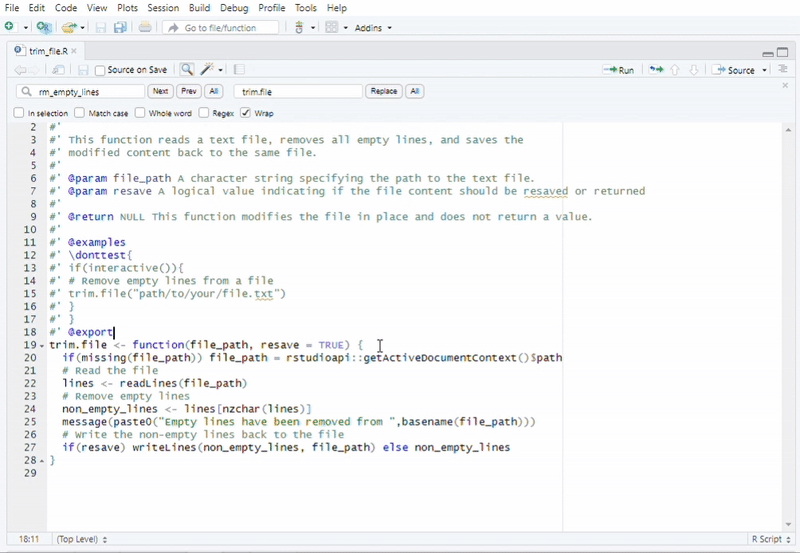
#Recommended to use "Addins" in RStudio
#Function call: Remove Empty Lines from a File
trim.file("path/to/your/file.txt")
#Detect outliers in data by IQR and Zscore methods
x <- c(1, 2, 3, 4, 100)
detect_outlier(x, summary = TRUE)
detect_outlier(z, method = "iqr", multiplier = 3)
detect_outlier(y, method = "zscore", z_threshold = 2.5)
#apply the yesNoBool to convert between boolean
#input type is "data.frame"
usedata <- data.frame(ID = number(32))
usedata #view the dataset
usedata$yess = rep(c("yes","n","no","YES","No","NO","yES","Y"),4) #create a new column
usedata #view the modified dataset
#set all yess field as standardize boolean
yesNoBool(usedata,yess, type="bin") #set all as binary 1/0
yesNoBool(usedata,yess, type="log") #set all as logical TRUE/FALSE
#initialize one or more variables
print(g) # Error: object 'g' not found
init(g,h,i,o)
print(g) # g = NULL
print(h) # h = NULL
init(r,y,u,b,value = 5)
print(r) # r = 5
print(b) # b = 5
print(z) # Error: object 'z' not found
#add keys to a vector content for use in downstream processes
ver1 <- c("Test 1","Test 2","Test 3")
add_key(ver1)
for(i in ver1){
message(sprintf("%s is the key for this %s", i$key, i$value))
}
# Introducing the super variable
# store dataset that should not be altered
newSuperVar(mtdf, value = austres) # create a super variable
head(mtdf) # view it
mtdf.class # view the store class of the variable, it cannot be changed
# it means that when the super variable is edited, the new value MUST have the same class
# create and lock super variable by default
# extra security to prevent changing
newSuperVar(mtdf3, value = beaver1, lock = TRUE)
head(mtdf3) # view
mtdf3.round(1) # round to 1 decimal places
head(mtdf3) # view
mtdf3.signif(2) # round to 2 significant digits
head(mtdf3) # view
# Task: create a new super variable to store numbers
# edit the numbers from various scopes
newSuperVar(edtvec, value = number(5))
edtvec # view content of the vector
# edtvec.set(letters) #ERROR: Cannot set to value with different class than initial value
edtvec.set(number(20)) # set to new numbers
edtvec # view output
for (pu in 1:8) {
print(edtvec) # view output within loop
edtvec.set(number(pu)) # set to new numbers within for loop
}
lc <- lapply(1:8, function(pu) {
print(edtvec) # view output within loop
edtvec.set(number(pu)) # set to new numbers within lapply loop
})
# see that the above changed the super variable easily.
# local variable will not be altered by the loop
# example
bim <- 198
lc <- lapply(1:8, function(j) {
print(bim)
bim <- j # will not alter the value of bim in next round
})
#check if the entry is not integer
not.integer(45) #returns TRUE
not.integer(45.) #returns TRUE
not.integer(45L) #returns FALSE
not.null(45L) #returns TRUE
not.null(h<-NULL) #returns FALSE
#clear R environment, set directory and load data
#note: the code below also automatically loads the quickcode library so that all other functions within package can be used easily
quickcode::refresh()
quickcode::clean()
#or combine with setwd and source and load
quickcode::clean(
setwd = "/wd/",
source = c(
"file.R",
"file2.R"
),
load = c(
"data.RData",
"data2.RData"
)
)
#shorthand for not in vector
p1 <- 4
p2 <- c(1:10)
p1 %nin% p2
#add to a vector in one code
p1 <- c(6,7,8)
p2 <- c(1,2,3)
vector_push(p1,p2)
print(p1)
#add to a data frame in one code
p1 <- data.frame(ID=1:10,ID2=1:10)
p2 <- data.frame(ID=11:20,ID2=21:30)
data_push(p1,p2,"rows")
print(p1)
#remove from a vector in one code
p1 <- c(6,7,8,1,2,3)
vector_pop(p1)
print(p1)
#remove from a data frame in one code
p1 <- data.frame(ID=1:10,ID2=1:10,CD=11:20,BD=21:30)
data_pop(p1) #remove last row
print(p1)
data_pop(p1,5) #remove last 5 rows
print(p1)
#remove columns from a data frame in one code
p1 <- data.frame(ID=1:10,ID2=1:10,ID4=1:10,CD=11:20,BD=21:30)
data_pop(p1,which = "cols") #remove last column
print(p1)
data_pop(p1,2,which = "cols") #remove last 2 columns
print(p1)
data_pop(p1,1,which = "cols") #remove last 1 column and vectorise
print(p1)